introCSB
R shiny and markdown resources for learning Computational Systems Biology
R Resources for Learning Computational Systems Biology
On this page you will find a variety of resources for teaching/learning Computational Systems Biology that have been developed using the R statistical language. These resources include interactive modules that are generally applicable to learning the basics of systems biology modeling, as well as, coding examples for using R to develop and analyze various types of systems biology models. The coding examples also include R shiny examples, which provide a template for developing interactive interfaces for systems biology models. Source codes for all interfaces and coding examples are also available. An interactive slide deck, also developed with R shiny, that was presented at FOSBE 2022 in a session on Systems Biology Eucation are available here.
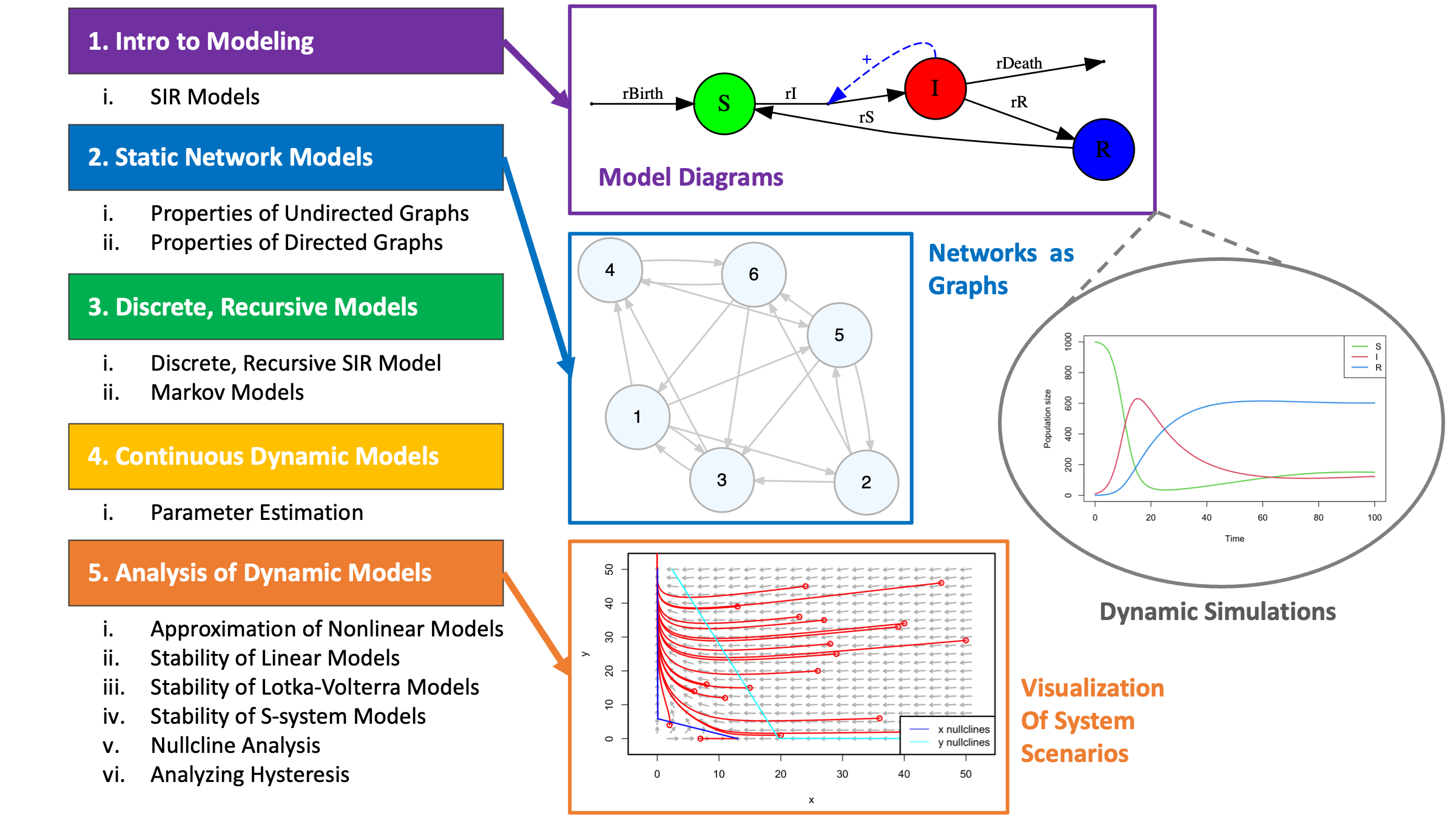 Schematic overview of resources in the introCSB repository.
Schematic overview of resources in the introCSB repository.
R Shiny Interactive Modules
This is a series of modules that are intended to provide simple interfaces to example systems models and to provide extra practice problems and are intended as a supplement the textbook: A First Course in Systems Biology . The textbook is not needed to be able to use these applications, but may be useful for further explanation.
- Intro to Modeling (SIR Models): Provides interfaces to selected examples from Chapter 2: Introduction to Mathematical Modeling of A First Course in Systems Biology. The expected outcomes of this module are (i) learn the parts of a model and (ii) practice predicting how perturbations to model inputs and parameters affect the model response. (Source)
- Static Network Models: Provides interfaces to example static network models in line with Chapter 3: Static Network Models of A First Course in Systems Biology. The expected outcomes of this module are (i) learn the basics of working with graphs; (ii) practice computing statistics based on graphs; (iii) practice using stoichiometric network models. (Source)
- Discrete, Recursive Models: Provides interfaces to example discrete and recursive models, including examples from Chapter 4: The Mathematics of Biological Systems of A First Course in Systems Biology. The expected outcomes of this module are (i) learn the basic principles of discrete, recursive models; (ii) explore examples of dynamic discrete recrusive models; (iii) practice generating Markov matrices. (Source)
- Continuous Dynamic Models: Provides interfaces to example models based on the canonical models described in pg. 102-105 of Chapter 4: The Mathematics of Biological Systems of A First Course in Systems Biology. The expected outcomes of this module are to learn the basic structure of (i) linear, (ii) Lotka-Voltera, (iii) mass action, and (iv) S-system models. (Source)
- Analysis of Dynamic Models: Provides interfaces to example models described in Chapter 4: The Mathematics of Biological Systems of A First Course in Systems Biology. The expected outcomes of this module are to learn the fundamentals of stability analysis including: (i) linearization of nonlinear models, (ii) eigenvalue analysis. (Source)
Interactive Coding Tutorials
These coding tutorials were developed using the learnr package and initial development was funded through a seed grant from the CACHE corporation:
- Introduction to Modeling Biological Systems: This tutorial introduces basic skills for performing systems-scale modeling of biological systems in R, including model diagrams and ODE based simulations. (Source)
R Coding Examples by Topic
Below is a list of links to HTML pages with coding examples for a range of topics related to developing and analysiing systems biology models. The examples cover developing systems diagrams and developing/analyzing ODE-based systems models.
- Intro to Modeling
- Properties of Undirected Graphs
- Properties of Directed Graphs
- Discrete, Recursive Models
- Markov Models
- Parameter Estimation
- Stability of Linear Models
- Approximation of Nonlinear Models
- Stability of Lotka-Volterra Models
- Stability of S-system Models
- Nullcline Analysis
- Analyzing Hysteresis
R shiny Examples
shiny is an R package that enables the efficient development of web-based applications. The following examples demonstrate how R shiny can be used for developing interactive interfaces for systems biology models.
- Interface for ODE based model: Simple example of how to use R shiny to develop interactive interfaces for ODE-based models. (Demo)
- Interface for ODE based model with report export: Advanced example of how to use R shiny to develop interactive interfaces for ODE-based models. Includes the ability to export analysis from the model using an R markdown derived report in PDF format. (Demo)
Miscellaneous R shiny Applications
In addition to the interactive modules and the R shiny examples above, here is a list of miscellaneous applications for dynamic systems modeling.
- Simple interfaces to example dynamic systems.
- Series applications related to stability analysis of linear systems.
- Practice problems for stability analysis of Lotka-Volterra (predator-prey) systems.
- Practice problems for nullcline analysis of Lotka-Volterra (predator-prey) systems.
- Practice problems for stability analysis of S-systems.
- Practice problems for building Markov matrices from directed graphs.
Citation: Voit, E.O.: A First Course in Systems Biology. Garland Science, New York, NY, 2017, 2nd edition